library(flowCore)
library(flowWorkspace)
library(openCyto)
library(Luciernaga)
library(flowSpectrum)
library(dplyr)
library(purrr)
library(stringr)
library(ggplot2)
path <- file.path("/media", "david", "David")
files <- list.files(path, pattern=".fcs", full.names=TRUE, recursive=FALSE)
#files <- files[str_detect(files, "12 ")]
MyCytoSet <- load_cytoset_from_fcs(files, transformation=FALSE, truncate_max_range = FALSE)
MyGatingSet <- GatingSet(MyCytoSet)
MyGates <- data.table::fread("/home/david/Documents/CytometryInR/data/QCBeadGates.csv")
MyGatingTemplate <- gatingTemplate(MyGates)
gt_gating(MyGatingTemplate, MyGatingSet)
#pData(MyGatingSet)
#plot(MyGatingSet)
#x <- MyGatingSet[1]
#subset <- "beads"
#sample.name <- "TUBENAME"
RCVfromFCS <- function(x, subset, sample.name){
Name <- keyword(x, sample.name)
Internal <- gs_pop_get_data(x, subset)
Internal <- exprs(Internal[[1]])
Internal <- data.frame(Internal, check.names=FALSE)
These <- colnames(Internal)
TheRCVs <- map(.x=These, .f=InternalRCV, data=Internal) |>
bind_cols()
TheRCVs <- TheRCVs |> mutate(Sample=Name) |>
relocate(Sample, .before=1)
return(TheRCVs)
}
#data <- Internal
#x <- These[1]
InternalRCV <- function(x, data){
Name <- x
TheCol <- data |> select(x)
TheCol <- TheCol[!is.na(TheCol)]
if(length(TheCol) == 0) return(NULL)
RCV <- mad(TheCol) / median(TheCol)
RCV <- data.frame(RCV)
colnames(RCV) <- Name
return(RCV)
}
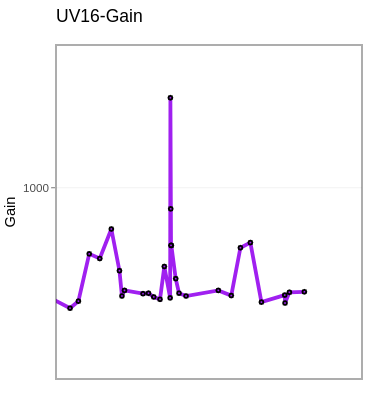
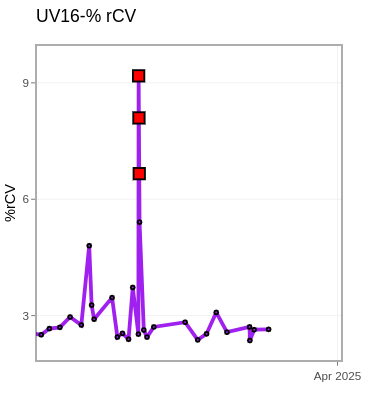
RCVs <- map(.x=MyGatingSet, .f=RCVfromFCS, subset="beads", sample.name="TUBENAME") |> bind_rows()
LastDetectors <- RCVs |> select(Sample, "UV16-A", "V16-A", "B14-A", "YG10-A", "R8-A")
LastDetectors[,2:6] <- LastDetectors[,2:6]*100
LastDetectors[,2:6] <- round(LastDetectors[,2:6], 2)
FileLocation <- system.file("extdata", package = "Luciernaga")
pattern = "AutofluorescentOverlaps.csv"
AFOverlap <- list.files(path=FileLocation, pattern=pattern,
full.names = TRUE)
AFOverlap_CSV <- read.csv(AFOverlap, check.names = FALSE)
#AFOverlap_CSV
#pData(SpectraData)
SpectraData <- gs_pop_get_data(MyGatingSet, "beads")
SpectraData <- flowWorkspace::cytoset_to_flowSet(SpectraData)
outpath <- file.path("media", "david", "Desktop")
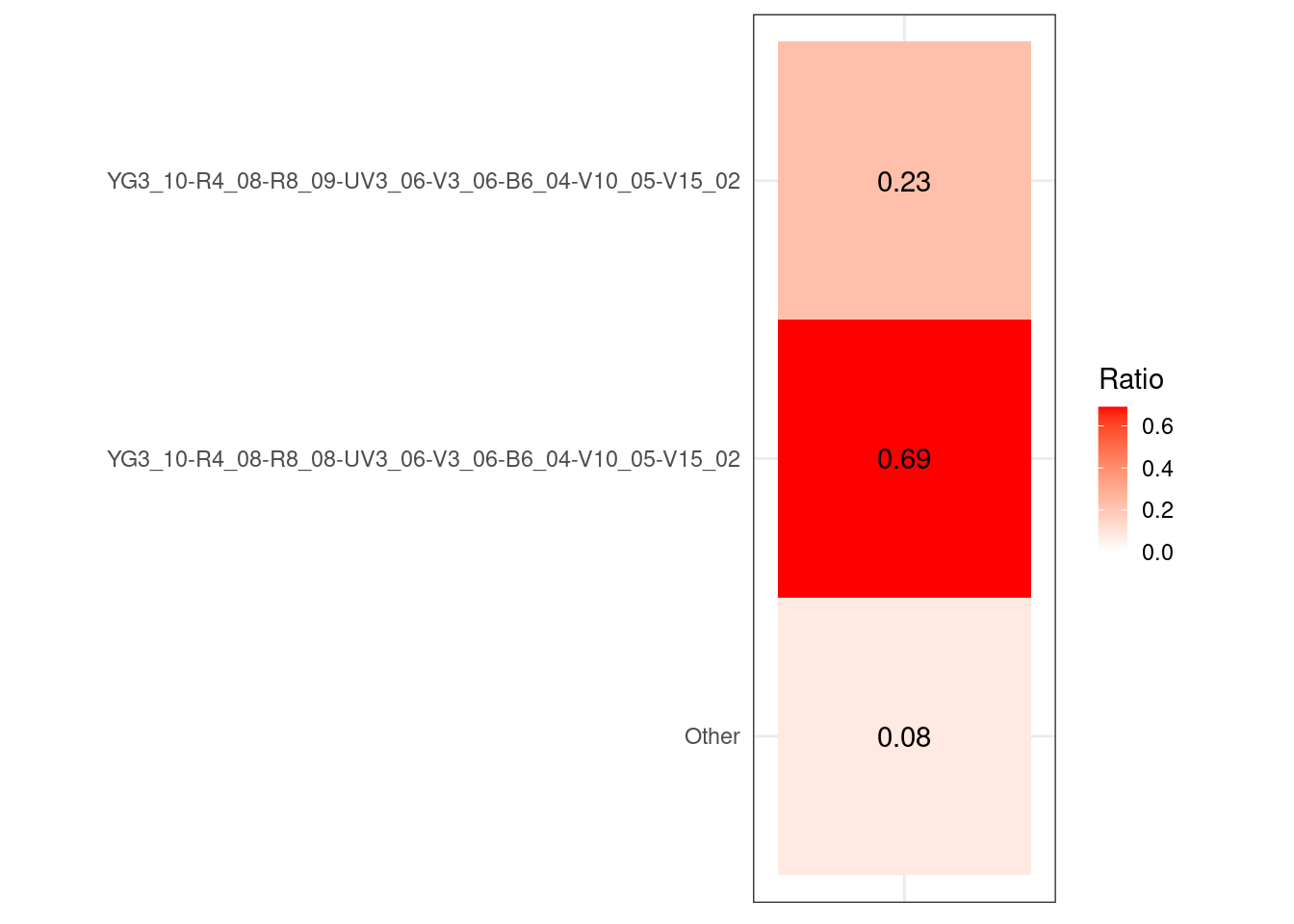
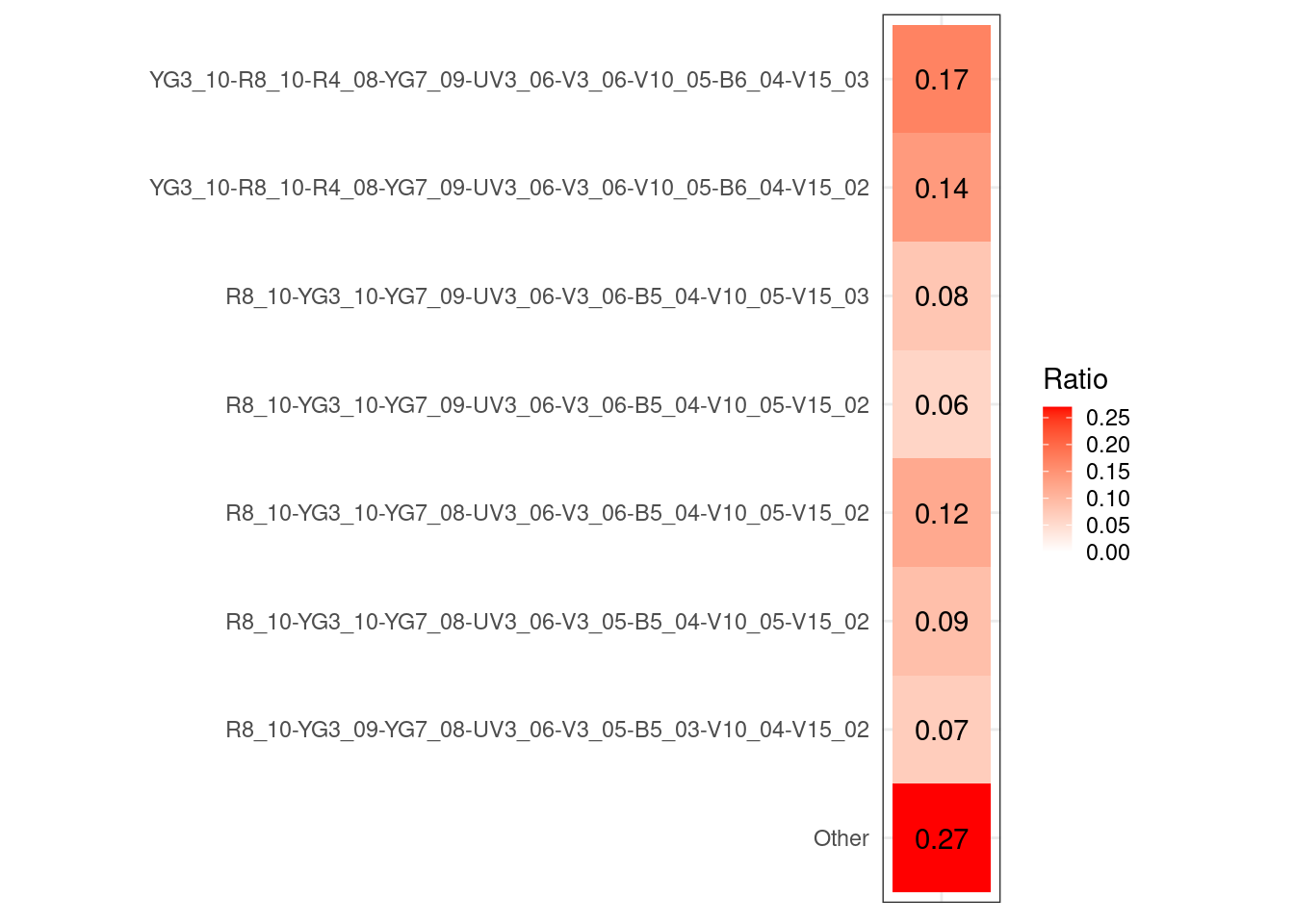
UnstainedSignature <- map(.x=MyGatingSet, .f=Luciernaga_QC,
subsets="beads",
removestrings=".fcs",
sample.name="TUBENAME",
unmixingcontroltype = "cells",
Unstained = TRUE,
ratiopopcutoff = 0.001,
Verbose = TRUE,
AFOverlap = AFOverlap,
stats = "median",
ExportType = "data",
SignatureReturnNow = FALSE,
outpath = outpath,
Increments=0.1, experiment="Lot2006",
condition.name="$DATE", SecondaryPeaks=8) |>
bind_rows()
#UnstainedSignature$Condition <- lubridate::dmy(UnstainedSignature$Condition)
#str(UnstainedSignature)
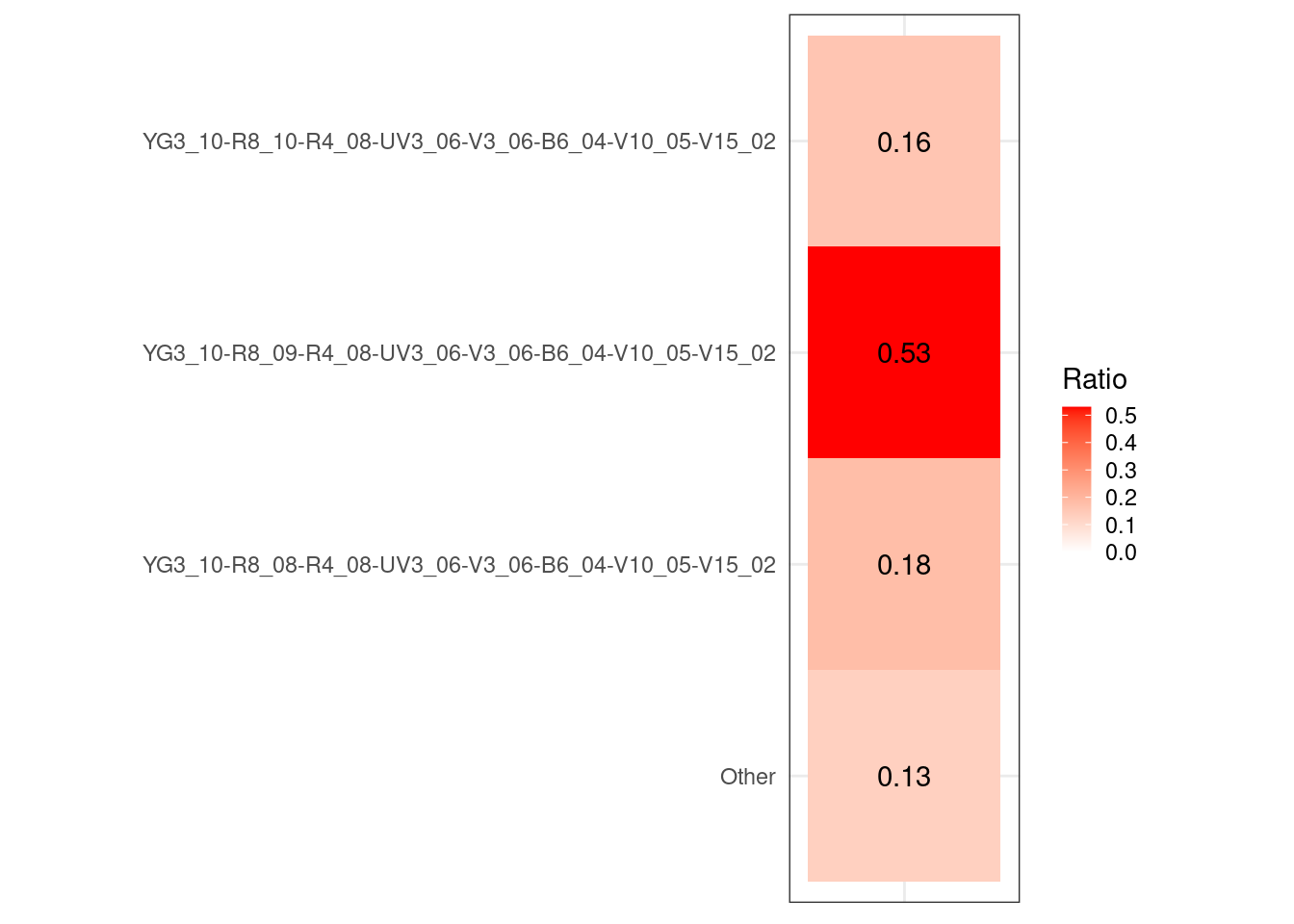
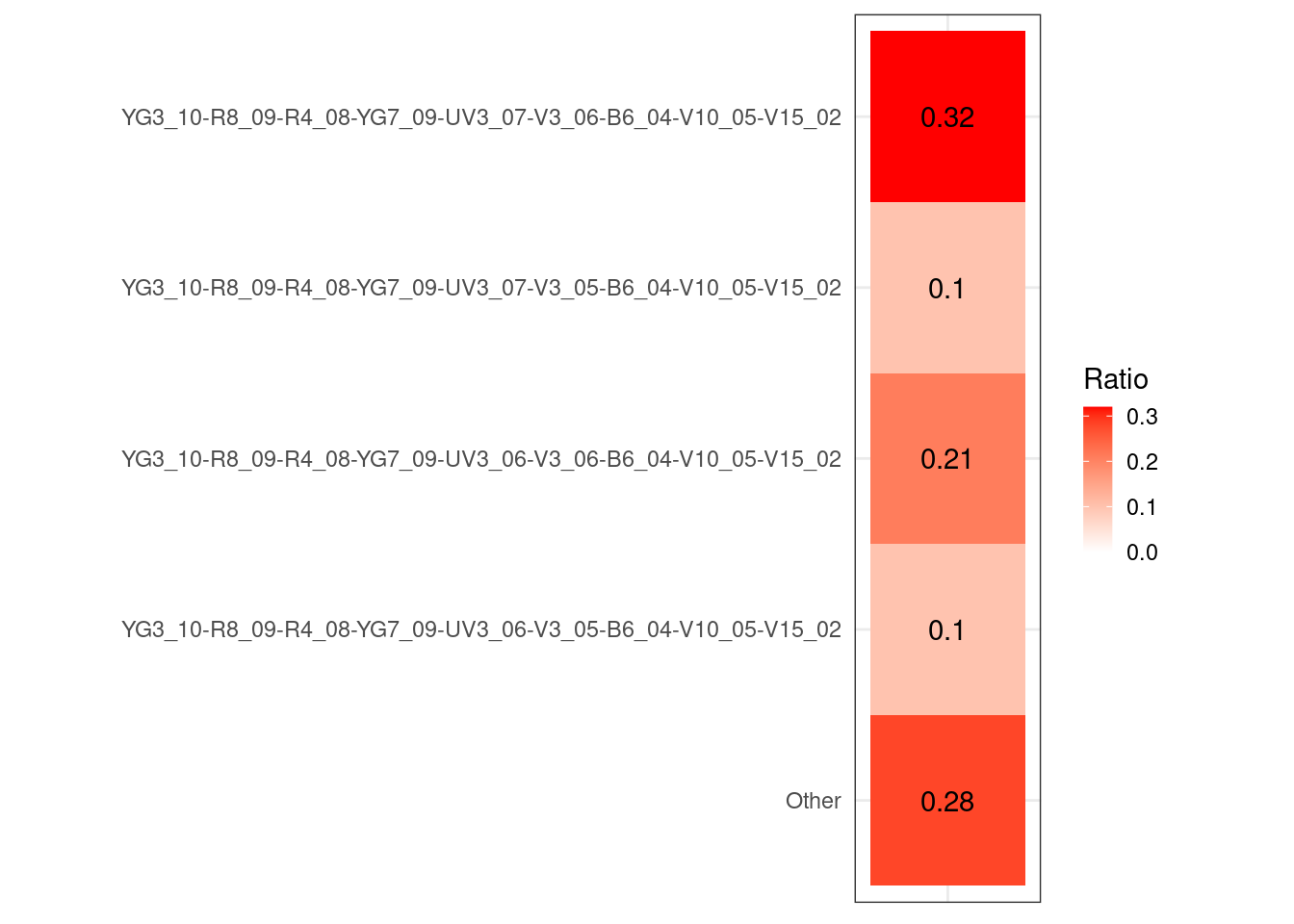
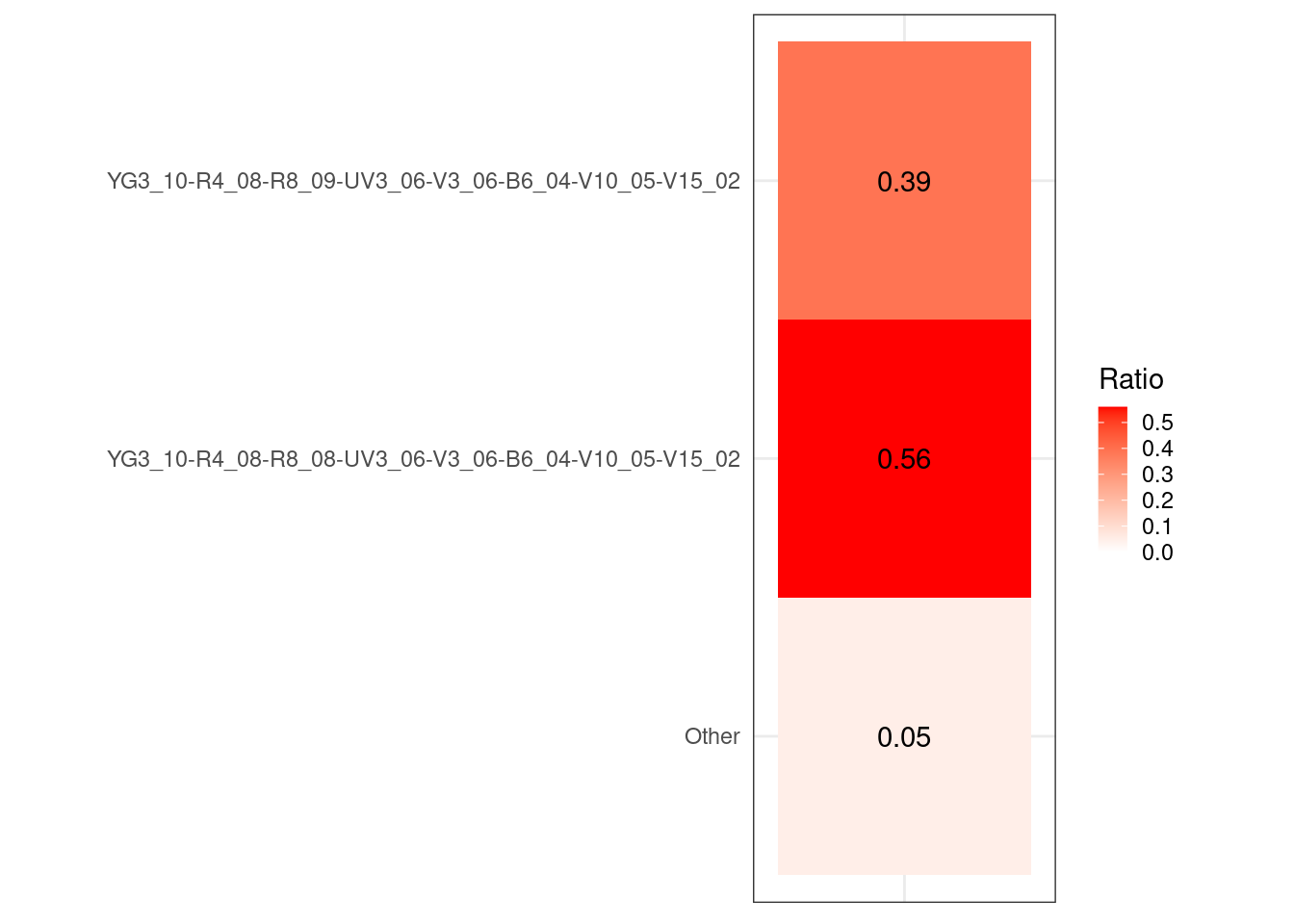
#table(UnstainedSignature$Cluster)